Bootstrapping
TODO
- link to resamplingBootALM
Install required packages
wants <- c("boot")
has <- wants %in% rownames(installed.packages())
if(any(!has)) install.packages(wants[!has])Confidence interval for \(\mu\)
Using package boot
set.seed(123)
muH0 <- 100
sdH0 <- 40
N <- 200
DV <- rnorm(N, muH0, sdH0)Function to calculate the mean and uncorrected variance (=plug-in estimator for the population variance) of a given replication.
getM <- function(orgDV, idx) {
n <- length(orgDV[idx])
bsM <- mean(orgDV[idx])
bsS2M <- (((n-1) / n) * var(orgDV[idx])) / n
c(bsM, bsS2M)
}
library(boot)
nR <- 999
(bsRes <- boot(DV, statistic=getM, R=nR))
ORDINARY NONPARAMETRIC BOOTSTRAP
Call:
boot(data = DV, statistic = getM, R = nR)
Bootstrap Statistics :
original bias std. error
t1* 99.657182 0.1252697500 2.5831186
t2* 7.080822 0.0008318941 0.7763612Various types of bootstrap confidence intervals
alpha <- 0.05
boot.ci(bsRes, conf=1-alpha, type=c("basic", "perc", "norm", "stud", "bca"))BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS
Based on 999 bootstrap replicates
CALL :
boot.ci(boot.out = bsRes, conf = 1 - alpha, type = c("basic",
"perc", "norm", "stud", "bca"))
Intervals :
Level Normal Basic Studentized
95% ( 94.47, 104.59 ) ( 94.59, 104.84 ) ( 94.77, 105.09 )
Level Percentile BCa
95% ( 94.48, 104.72 ) ( 94.20, 104.56 )
Calculations and Intervals on Original ScaleBootstrap distribution
For the \(t\) test statistic, compare the empirical distribution from the bootstrap replicates against the theoretical \(t_{n-1}\) distribtion.
res <- replicate(nR, getM(DV, sample(seq(along=DV), replace=TRUE)))
Mstar <- res[1, ]
SMstar <- sqrt(res[2, ])
tStar <- (Mstar-mean(DV)) / SMstarplot(tStar, ecdf(tStar)(tStar), col="gray60", pch=1, xlab="t* bzw. t",
ylab="P(T <= t)", main="t*: cumulative rel. frequency and t CDF")
curve(pt(x, N-1), lwd=2, add=TRUE)
legend(x="topleft", lty=c(NA, 1), pch=c(1, NA), lwd=c(2, 2),
col=c("gray60", "black"), legend=c("t*", "t"))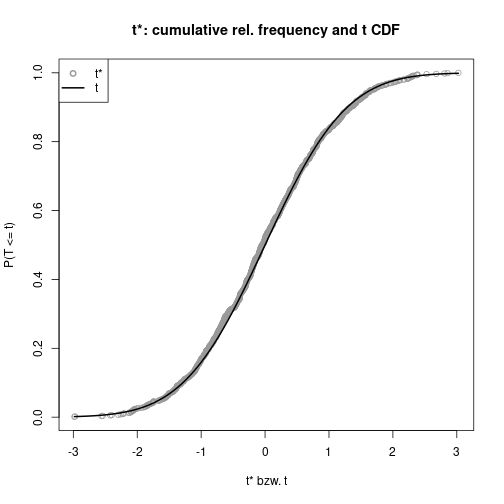
Detailed information about bootstrap samples
boot.array(boot(...), indices=TRUE) gives detailed information about the selected indices for each bootstrap replication. If the sample has \(n\) observations, and there are \(R\) replications, the result is an \((R \times n)\)-matrix with one row for each replication and one column for each observation.
bootIdx <- boot.array(bsRes, indices=TRUE)
# replications 1-3: first 10 selected indices in each replication
bootIdx[1:3, 1:10] [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
[1,] 198 48 130 122 114 89 100 178 103 144
[2,] 28 47 181 101 164 69 20 111 35 46
[3,] 182 47 147 101 152 104 58 118 52 33# selected indices in the first replication
repl1Idx <- bootIdx[1, ]
# selected values in the first replication
repl1DV <- DV[repl1Idx]
head(repl1DV, n=5) [1] 49.94915 81.33379 97.14768 62.10102 97.77752Detach (automatically) loaded packages (if possible)
try(detach(package:boot))Get the article source from GitHub
R markdown - markdown - R code - all posts