Diagram types with the ggplot2 package
Install required packages
ggplot2, ggbeeswarm, cowplot, patchwork
wants <- c("ggplot2", "ggbeeswarm", "cowplot", "patchwork")
has <- wants %in% rownames(installed.packages())
if(any(!has)) install.packages(wants[!has])Simulate data
Njk <- 50
P <- 4
Q <- 2
IQ <- rnorm(P*Q*Njk, mean=100, sd=15)
height <- rnorm(P*Q*Njk, mean=175, sd=7)
rating <- factor(sample(LETTERS[1:3], Njk*P*Q, replace=TRUE))
sex <- factor(rep(c("f", "m"), times=P*Njk))
group <- factor(rep(c("control", "placebo", "treatA", "treatB"), each=Q*Njk))
sgComb <- interaction(sex, group)
mood <- round(rnorm(P*Q*Njk, mean=c(85, 80, 110, 90, 130, 100, 93, 121)[sgComb], sd=25))
myDf <- data.frame(sex, group, sgComb, IQ, height, rating, mood)Diagram types
Scatterplot
Line plot
group=TRUE is necessary here
group mood
1 control 86.79
2 placebo 104.29
3 treatA 114.53
4 treatB 105.14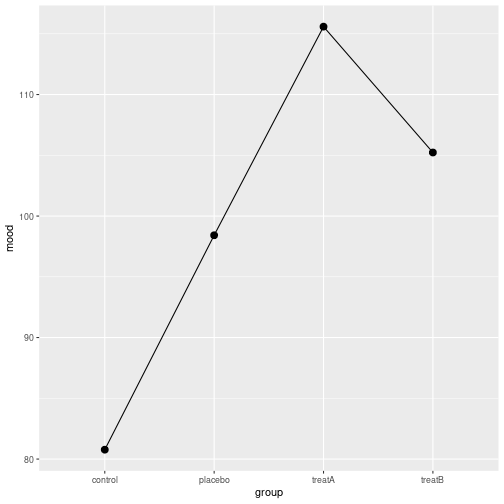
group mood
1 control 86.79
2 placebo 104.29
3 treatA 114.53
4 treatB 105.14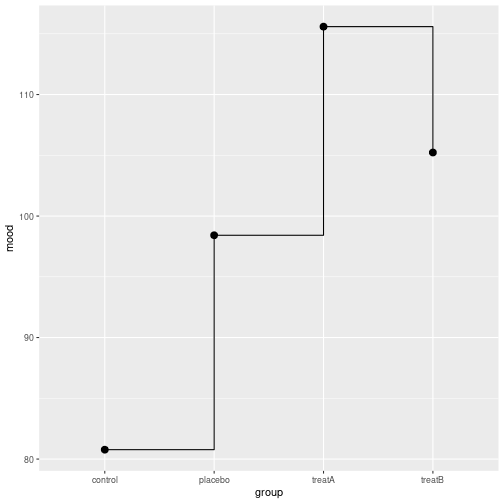
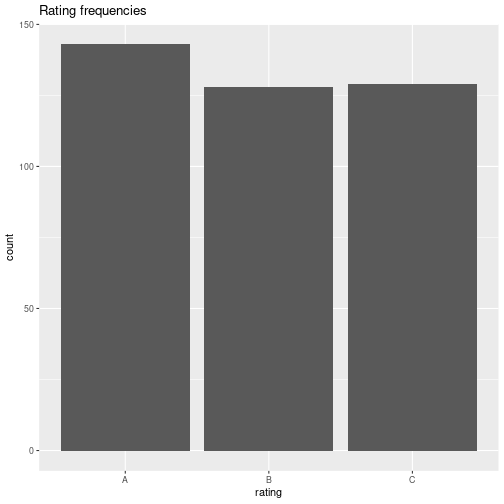
Barplot
Absolute frequencies
Relative frequencies
ggplot(myDf, aes(x=rating)) +
geom_bar(stat="count",
aes(y=(..count..) / sum(..count..))) +
ggtitle("Relative rating frequencies")
Histogram
Absolute frequencies
ggplot(myDf, aes(x=IQ)) +
geom_histogram(bins=20) +
ggtitle("Histogram IQ by group") +
theme(legend.position="none")
Density with nonparametric kernel density estimator
ggplot(myDf, aes(x=mood)) +
geom_histogram(aes(y=..density..)) +
ggtitle("Histogram mood by group") +
geom_density(color="darkgrey", fill="grey", alpha=0.6)
Boxplot
Add a geom_beeswarm() layer to show raw data.
library(ggbeeswarm)
ggplot(myDf, aes(x=sex, y=height)) +
geom_boxplot(outlier.shape=NULL) +
geom_beeswarm(alpha=0.5) +
ggtitle("Height by sex") +
theme(legend.position="none")
Q-Q plot
Against normal distribution
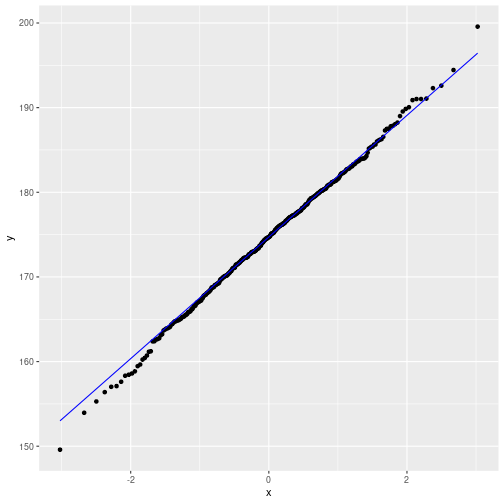
Against pre-specified distribution (here: t)
parL <- list(df=10, ncp=0)
ggplot(myDf, aes(sample=height)) +
geom_qq(distribution=qt, dparams=parL) +
geom_qq_line(color="blue", distribution=qt, dparams=parL)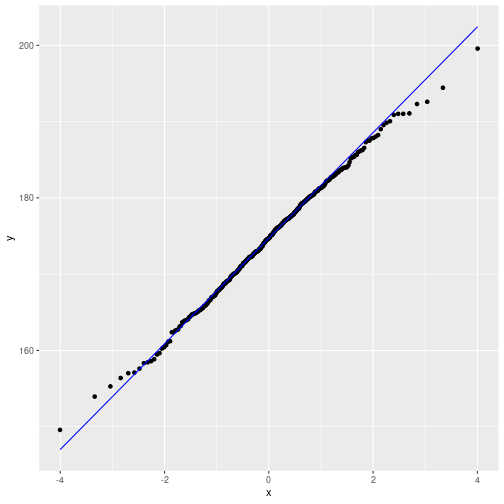
Compare quantiles of two variables
probs <- ppoints(50)
qDat <- data.frame(qmood=quantile(myDf$mood, probs),
qheight=quantile(myDf$height, probs))
ggplot(qDat, aes(x=qmood, y=qheight)) +
geom_point() +
xlab("Quantiles mood") +
ylab("Quantiles height")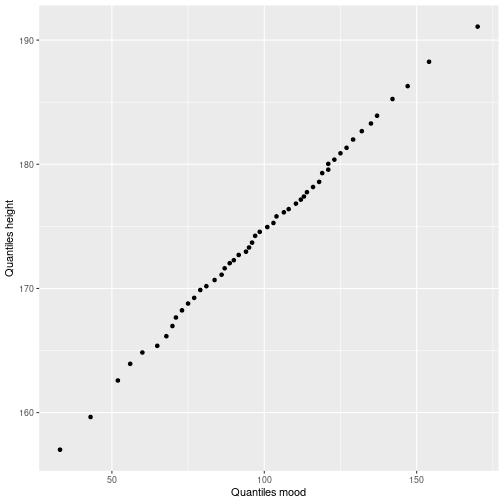
Multi-panel plots
Split the diagram into panels by column using facet_grid().
ggplot(myDf, aes(x=rating, y=IQ)) +
geom_point(size=3) +
facet_grid(. ~ group) +
ggtitle("IQ ~ height split by sex and group") +
guides(shape="none")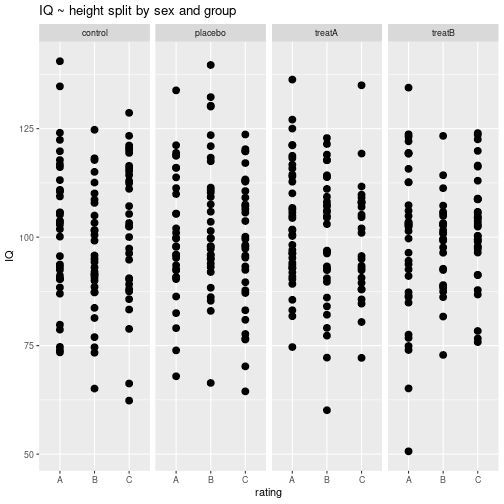
Split the diagram into matrix of panels using facet_wrap(). Option scales="free_y" produces separate y-axis for each panel.
ggplot(myDf, aes(x=rating, y=IQ)) +
geom_point(size=3) +
facet_wrap(. ~ group, scales="free_y") +
ggtitle("IQ ~ height split by sex and group") +
guides(shape="none")
Additional diagram elements
Add error bars
groupN <- as.data.frame(xtabs(~ sex + group, data=myDf))
groupM <- aggregate(mood ~ sex + group, data=myDf, FUN=mean)
groupSD <- aggregate(mood ~ sex + group, data=myDf, FUN=sd)
groupMSD <- merge(groupM, groupSD, by=c("sex", "group"), suffixes=c(".M", ".SD"))
groupMSDN <- merge(groupMSD, groupN, by=c("sex", "group"))
(groupMSDN <- transform(groupMSDN,
SEMlo=mood.M - mood.SD/sqrt(Freq),
SEMup=mood.M + mood.SD/sqrt(Freq))) sex group mood.M mood.SD Freq SEMlo SEMup
1 f control 90.16 26.10916 50 86.46761 93.85239
2 f placebo 115.32 22.71064 50 112.10823 118.53177
3 f treatA 125.52 27.40791 50 121.64394 129.39606
4 f treatB 91.36 25.60035 50 87.73956 94.98044
5 m control 83.42 25.34826 50 79.83521 87.00479
6 m placebo 93.26 23.49452 50 89.93737 96.58263
7 m treatA 103.54 23.58416 50 100.20470 106.87530
8 m treatB 118.92 22.67772 50 115.71289 122.12711ggplot(groupMSDN, aes(x=group, y=mood.M, ymin=SEMlo, ymax=SEMup,
color=sex, shape=sex, group=sex)) +
geom_point() +
geom_line() +
geom_linerange()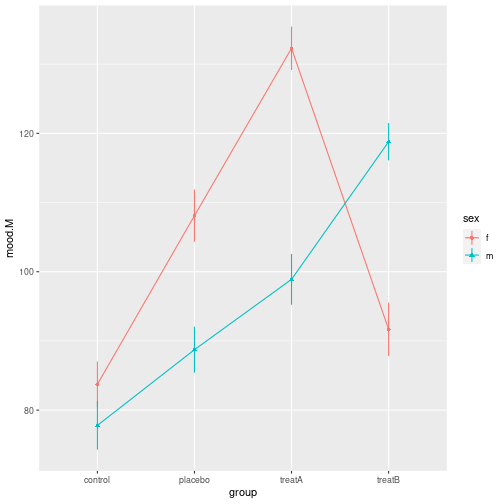
geom_hline()for a horizontal linegeom_vline()for a vertical linegeom_smooth()for a regression line including SEsgeom_text()for text from a variableggtitle()for a diagram titleannotate()for text given directly as an argument
ggplot(myDf, aes(x=height, y=IQ)) +
geom_hline(aes(yintercept=100), linetype=2) +
geom_vline(aes(xintercept=180), linetype=2) +
geom_point(size=3) +
geom_smooth(method=lm, se=TRUE, size=1.2, fullrange=TRUE) +
facet_grid(sex ~ group) +
ggtitle("IQ ~ height split by sex and group") +
theme(legend.position="none") +
geom_text(aes(x=190, y=70, label=sgComb)) +
annotate("text", x=165, y=130, label="annotation")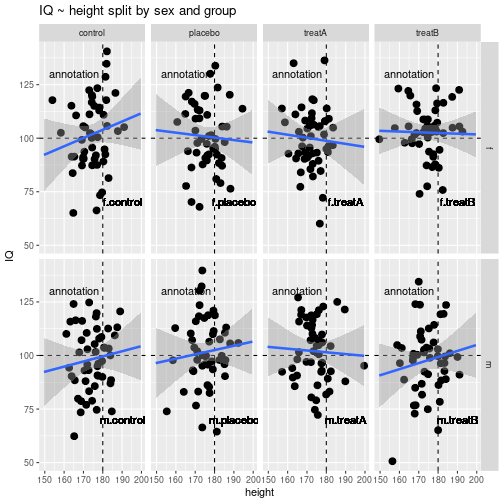
Put different plots on the same page
Using package cowplot
library(cowplot)
p1 <- ggplot(myDf, aes(x=height, y=IQ)) +
geom_point()
p2 <- ggplot(myDf, aes(x=rating)) +
geom_bar(stat="count", position=position_stack()) +
ggtitle("Rating frequencies by sex and group")
plot_grid(p1, p2, ncol=1, align="v")
Using package patchwork
Save ggplot2 diagrams to file
Graphics file format is determined from file ending.
p <- ggplot(myDf, aes(x=height, y=IQ)) +
geom_point(size=3)
ggsave(p, file="diag_ggplot.pdf", width=9, height=6, units="in")
ggsave(p, file="diag_ggplot.png", width=200, height=150, units="mm", dpi=300)Further online resources
- See Cookbook for R:
ggplot2diagrams for many detailed examples ofggplot2diagrams. - Package
esquisseprovides an interactive Shiny app to create and customize ggplot2 diagrams.
Detach (automatically) loaded packages (if possible)
try(detach(package:cowplot))
try(detach(package:patchwork))
try(detach(package:ggbeeswarm))
try(detach(package:ggplot2))Get the article source from GitHub
R markdown - markdown - R code - all posts