Regression diagnostics
Regression diagnostics
TODO
- link to regression, regressionLogistic
Install required packages
car, lmtest, mvoutlier, perturb, robustbase, tseries
wants <- c("car", "lmtest", "mvoutlier", "perturb", "robustbase", "tseries")
has <- wants %in% rownames(installed.packages())
if(any(!has)) install.packages(wants[!has])Extreme values and outliers
Univariate assessment of outliers
set.seed(123)
N <- 100
X1 <- rnorm(N, 175, 7)
X2 <- rnorm(N, 30, 8)
X3 <- 0.3*X1 - 0.2*X2 + rnorm(N, 0, 5)
Y <- 0.5*X1 - 0.3*X2 - 0.4*X3 + 10 + rnorm(N, 0, 5)
dfRegr <- data.frame(X1, X2, X3, Y)library(robustbase)
xyMat <- data.matrix(dfRegr)
robXY <- covMcd(xyMat)
XYz <- scale(xyMat, center=robXY$center, scale=sqrt(diag(robXY$cov)))
summary(XYz) X1 X2 X3
Min. :-2.53818 Min. :-1.96554 Min. :-2.4625
1st Qu.:-0.63344 1st Qu.:-0.68918 1st Qu.:-0.5474
Median :-0.05045 Median :-0.10278 Median : 0.1471
Mean :-0.02039 Mean : 0.01779 Mean : 0.1410
3rd Qu.: 0.61065 3rd Qu.: 0.60431 3rd Qu.: 0.7874
Max. : 2.17984 Max. : 3.43113 Max. : 2.9812
Y
Min. :-2.45112
1st Qu.:-0.61660
Median :-0.04010
Mean :-0.04869
3rd Qu.: 0.60444
Max. : 2.45654 Multivariate assessment of outliers
Mahalanobis distance with robust estimate for the covariance matrix
mahaSq <- mahalanobis(xyMat, center=robXY$center, cov=robXY$cov)
summary(sqrt(mahaSq)) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.6179 1.3450 1.7460 1.8440 2.2510 3.9330 Nonparametric multivariate outlier detection with package mvoutlier
library(mvoutlier)
aqRes <- aq.plot(xyMat)Projection to the first and second robust principal components.
Proportion of total variation (explained variance): 0.63336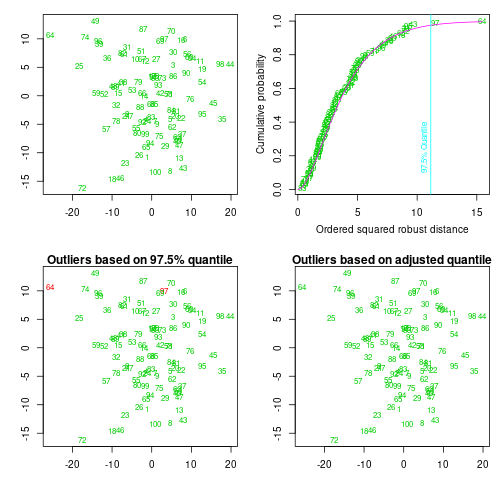
Where any outliers found?
which(aqRes$outliers)integer(0)Leverage and influence
fit <- lm(Y ~ X1 + X2 + X3, data=dfRegr)
h <- hatvalues(fit)
summary(h) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.01080 0.02190 0.03474 0.04000 0.05172 0.15250 cooksDst <- cooks.distance(fit)
summary(cooksDst) Min. 1st Qu. Median Mean 3rd Qu. Max.
8.300e-07 8.613e-04 3.650e-03 1.029e-02 1.362e-02 6.498e-02 inflRes <- influence.measures(fit)
summary(inflRes)Potentially influential observations of
lm(formula = Y ~ X1 + X2 + X3, data = dfRegr) :
dfb.1_ dfb.X1 dfb.X2 dfb.X3 dffit cov.r cook.d hat
44 -0.04 0.05 -0.03 -0.02 0.06 1.15_* 0.00 0.09
59 0.04 -0.04 -0.25 0.07 -0.38 0.83_* 0.03 0.02
60 0.04 0.15 -0.20 -0.45 0.52 0.84_* 0.06 0.04
64 0.05 -0.13 0.37 0.10 0.40 1.19_* 0.04 0.15_*
71 0.15 -0.20 0.02 0.17 0.34 0.84_* 0.03 0.02
74 0.05 -0.02 0.11 -0.12 0.21 1.14_* 0.01 0.10
95 0.04 -0.02 0.02 -0.07 -0.10 1.15_* 0.00 0.09
97 0.16 -0.10 -0.09 -0.13 -0.20 1.16_* 0.01 0.12 library(car)
influenceIndexPlot(fit)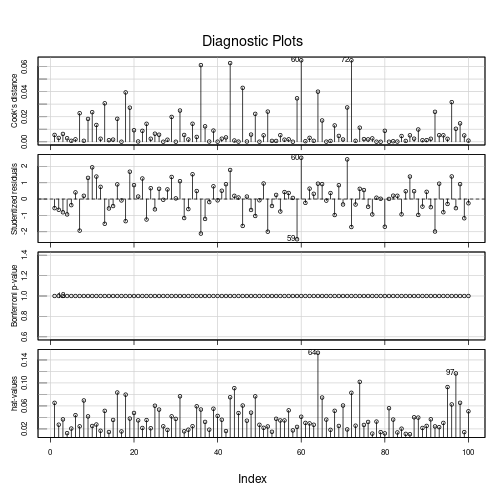
Checking model assumptions using residuals
Estnd <- rstandard(fit)
Estud <- rstudent(fit)Normality assumption
par(mar=c(5, 4.5, 4, 2)+0.1)
hist(Estud, main="Histogram studentized residals", breaks="FD", freq=FALSE)
curve(dnorm(x, mean=0, sd=1), col="red", lwd=2, add=TRUE)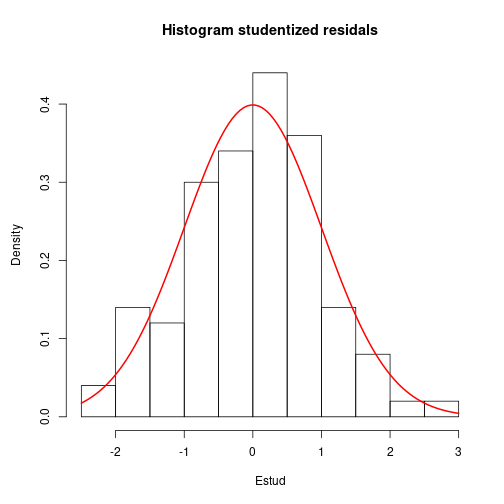
qqPlot(Estud, distribution="norm", pch=20, main="QQ-Plot studentized residuals")
qqline(Estud, col="red", lwd=2)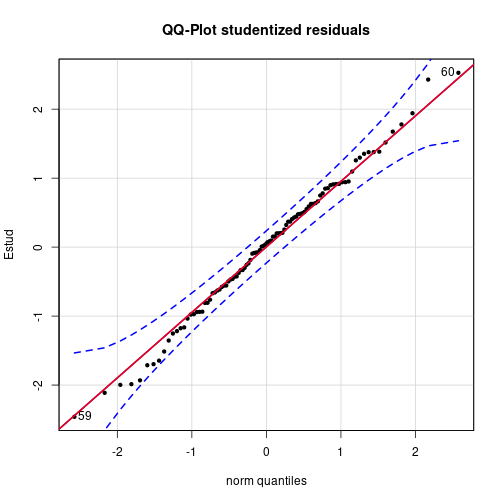
shapiro.test(Estud)
Shapiro-Wilk normality test
data: Estud
W = 0.9943, p-value = 0.9535Independence and homoscedasticity assumption
Spread-level plot
library(car)
spreadLevelPlot(fit, pch=20)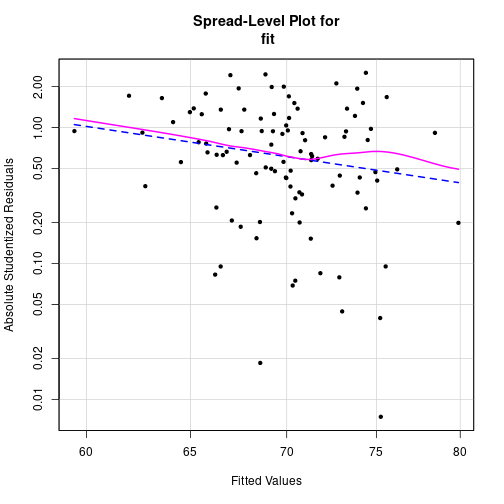
Suggested power transformation: 4.32355 Durbin-Watson-test for autocorrelation
library(car)
durbinWatsonTest(fit) lag Autocorrelation D-W Statistic p-value
1 -0.08825048 2.172797 0.36
Alternative hypothesis: rho != 0Statistical tests for heterocedasticity
Breusch-Pagan-Test
library(lmtest)
bptest(fit)
studentized Breusch-Pagan test
data: fit
BP = 1.9213, df = 3, p-value = 0.5889Score-test for non-constant error variance
library(car)
ncvTest(fit)Non-constant Variance Score Test
Variance formula: ~ fitted.values
Chisquare = 0.548752 Df = 1 p = 0.4588281 Linearity assumption
White-test
library(tseries)
white.test(dfRegr$X1, dfRegr$Y)
White Neural Network Test
data: dfRegr$X1 and dfRegr$Y
X-squared = 1.9818, df = 2, p-value = 0.3712white.test(dfRegr$X2, dfRegr$Y)
white.test(dfRegr$X3, dfRegr$Y)
# not runResponse transformations
lamObj <- powerTransform(fit, family="bcPower")
(lambda <- coef(lamObj)) Y1
1.280836 library(car)
yTrans <- bcPower(dfRegr$Y, lambda)Multicollinearity
Pairwise correlations between predictor variables
X <- data.matrix(subset(dfRegr, select=c("X1", "X2", "X3")))
(Rx <- cor(X)) X1 X2 X3
X1 1.00000000 -0.04953215 0.2700393
X2 -0.04953215 1.00000000 -0.2929049
X3 0.27003928 -0.29290486 1.0000000Variance inflation factor
library(car)
vif(fit) X1 X2 X3
1.079770 1.094974 1.178203 Condition indexes
\(\kappa\)
fitScl <- lm(scale(Y) ~ scale(X1) + scale(X2) + scale(X3), data=dfRegr)
kappa(fitScl, exact=TRUE)[1] 1.508749library(perturb)
colldiag(fit, scale=TRUE, center=FALSE)Condition
Index Variance Decomposition Proportions
intercept X1 X2 X3
1 1.000 0.000 0.000 0.004 0.001
2 8.371 0.001 0.001 0.781 0.029
3 26.110 0.046 0.040 0.208 0.964
4 78.331 0.953 0.959 0.008 0.006Using package perturb
pRes <- with(dfRegr, perturb(fit, pvars=c("X1", "X2", "X3"), prange=c(1, 1, 1)))Error in get(pvars): Objekt 'X1' nicht gefundensummary(pRes)Error in summary(pRes): Objekt 'pRes' nicht gefundenDetach (automatically) loaded packages (if possible)
try(detach(package:tseries))
try(detach(package:lmtest))
try(detach(package:zoo))
try(detach(package:perturb))
try(detach(package:mvoutlier))
try(detach(package:robustbase))
try(detach(package:sgeostat))
try(detach(package:car))Get the article source from GitHub
R markdown - markdown - R code - all posts