Analysis of covariance (ANCOVA)
TODO
- link to anovaSStypes
Install required packages
wants <- c("car", "effects", "multcomp")
has <- wants %in% rownames(installed.packages())
if(any(!has)) install.packages(wants[!has])Test the effects of group membership and of covariate
Visually assess the data
SSRIpre <- c(18, 16, 16, 15, 14, 20, 14, 21, 25, 11)
SSRIpost <- c(12, 0, 10, 9, 0, 11, 2, 4, 15, 10)
PlacPre <- c(18, 16, 15, 14, 20, 25, 11, 25, 11, 22)
PlacPost <- c(11, 4, 19, 15, 3, 14, 10, 16, 10, 20)
WLpre <- c(15, 19, 10, 29, 24, 15, 9, 18, 22, 13)
WLpost <- c(17, 25, 10, 22, 23, 10, 2, 10, 14, 7)P <- 3
Nj <- rep(length(SSRIpre), times=P)
dfAnc <- data.frame(IV=factor(rep(1:P, Nj), labels=c("SSRI", "Placebo", "WL")),
DVpre=c(SSRIpre, PlacPre, WLpre),
DVpost=c(SSRIpost, PlacPost, WLpost))plot(DVpre ~ IV, data=dfAnc, main="Pre-scores per group")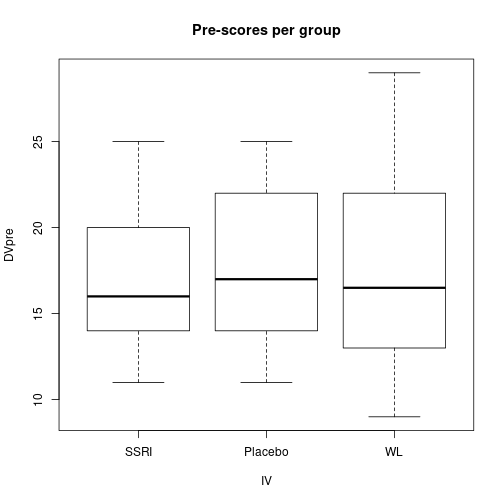
plot(DVpost ~ IV, data=dfAnc, main="Post-Scores per group")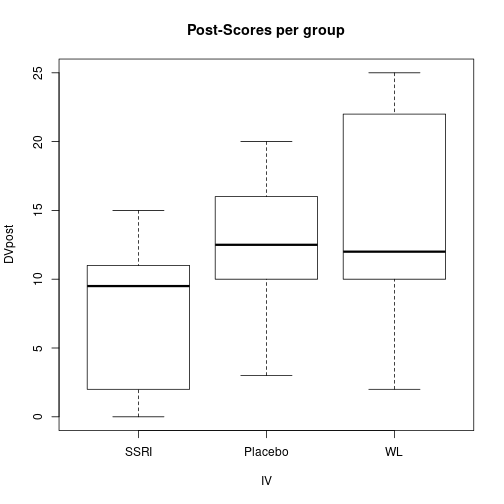
Type I sum of squares
fitFull <- lm(DVpost ~ IV + DVpre, data=dfAnc)
fitGrp <- lm(DVpost ~ IV, data=dfAnc)
fitRegr <- lm(DVpost ~ DVpre, data=dfAnc)anova(fitFull)Analysis of Variance Table
Response: DVpost
Df Sum Sq Mean Sq F value Pr(>F)
IV 2 240.47 120.23 4.1332 0.027629 *
DVpre 1 313.37 313.37 10.7723 0.002937 **
Residuals 26 756.33 29.09
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Type II/III sum of squares
Since no interaction is present in the model, SS type II and III are equivalent here.
Using Anova() from package car
library(car) # for Anova()
fitFiii <- lm(DVpost ~ IV + DVpre,
contrasts=list(IV=contr.sum), data=dfAnc)
Anova(fitFiii, type="III")Anova Table (Type III tests)
Response: DVpost
Sum Sq Df F value Pr(>F)
(Intercept) 0.00 1 0.0001 0.991035
IV 217.15 2 3.7324 0.037584 *
DVpre 313.37 1 10.7723 0.002937 **
Residuals 756.33 26
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Using model comparisons for SS type II
anova(fitRegr, fitFull)Analysis of Variance Table
Model 1: DVpost ~ DVpre
Model 2: DVpost ~ IV + DVpre
Res.Df RSS Df Sum of Sq F Pr(>F)
1 28 973.48
2 26 756.33 2 217.15 3.7324 0.03758 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(fitGrp, fitFull)Analysis of Variance Table
Model 1: DVpost ~ IV
Model 2: DVpost ~ IV + DVpre
Res.Df RSS Df Sum of Sq F Pr(>F)
1 27 1069.70
2 26 756.33 1 313.37 10.772 0.002937 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Test individual regression coefficients
(sumRes <- summary(fitFull))
Call:
lm(formula = DVpost ~ IV + DVpre, data = dfAnc)
Residuals:
Min 1Q Median 3Q Max
-10.6842 -3.9615 0.6448 3.8773 9.9675
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -3.6704 3.7525 -0.978 0.33703
IVPlacebo 4.4483 2.4160 1.841 0.07703 .
IVWL 6.4419 2.4133 2.669 0.01292 *
DVpre 0.6453 0.1966 3.282 0.00294 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 5.393 on 26 degrees of freedom
Multiple R-squared: 0.4227, Adjusted R-squared: 0.3561
F-statistic: 6.346 on 3 and 26 DF, p-value: 0.002252confint(fitFull) 2.5 % 97.5 %
(Intercept) -11.3836718 4.042941
IVPlacebo -0.5178144 9.414373
IVWL 1.4812101 11.402537
DVpre 0.2411673 1.049464Vsisualize ANCOVA coefficients
coeffs <- coef(sumRes)
iCeptSSRI <- coeffs[1, 1]
iCeptPlac <- coeffs[2, 1] + iCeptSSRI
iCeptWL <- coeffs[3, 1] + iCeptSSRI
slopeAll <- coeffs[4, 1]xLims <- c(0, max(dfAnc$DVpre))
yLims <- c(min(iCeptSSRI, iCeptPlac, iCeptWL), max(dfAnc$DVpost))
plot(DVpost ~ DVpre, data=dfAnc, xlim=xLims, ylim=yLims,
pch=rep(c(3, 17, 19), Nj), col=rep(c("red", "green", "blue"), Nj),
main="Data and group-wise regression lines")
legend(x="topleft", legend=levels(dfAnc$IV), pch=c(3, 17, 19),
col=c("red", "green", "blue"))
abline(iCeptSSRI, slopeAll, col="red")
abline(iCeptPlac, slopeAll, col="green")
abline(iCeptWL, slopeAll, col="blue")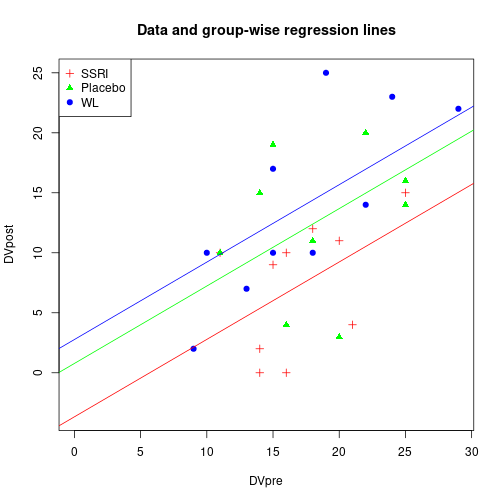
Effect size estimate
\(\hat{\omega}^{2}\) for the group effect
Using SS type II
anRes <- anova(fitRegr, fitFull)
dfGrp <- anRes[2, "Df"]
dfE <- anRes[2, "Res.Df"]
MSgrp <- anRes[2, "Sum of Sq"] / dfGrp
MSE <- anRes[2, "RSS"] / dfE
SST <- sum(anova(fitFull)[ , "Sum Sq"])
(omegaSqHat <- dfGrp*(MSgrp - MSE) / (SST + MSE))[1] 0.1187001Planned comparisons between groups
Adjusted group means
aovAncova <- aov(DVpost ~ IV + DVpre, data=dfAnc)
library(effects) # for effect()
YMjAdj <- effect("IV", aovAncova)
summary(YMjAdj)
IV effect
IV
SSRI Placebo WL
7.536616 11.984895 13.978489
Lower 95 Percent Confidence Limits
IV
SSRI Placebo WL
4.027629 8.476452 10.472608
Upper 95 Percent Confidence Limits
IV
SSRI Placebo WL
11.04560 15.49334 17.48437 Planned comparisons
cMat <- rbind("SSRI-Placebo" = c(-1, 1, 0),
"SSRI-WL" = c(-1, 0, 1),
"SSRI-0.5(P+WL)"= c(-2, 1, 1))library(multcomp) # for glht()
aovAncova <- aov(DVpost ~ IV + DVpre, data=dfAnc)
summary(glht(aovAncova, linfct=mcp(IV=cMat), alternative="greater"),
test=adjusted("none"))
Simultaneous Tests for General Linear Hypotheses
Multiple Comparisons of Means: User-defined Contrasts
Fit: aov(formula = DVpost ~ IV + DVpre, data = dfAnc)
Linear Hypotheses:
Estimate Std. Error t value Pr(>t)
SSRI-Placebo <= 0 4.448 2.416 1.841 0.03852 *
SSRI-WL <= 0 6.442 2.413 2.669 0.00646 **
SSRI-0.5(P+WL) <= 0 10.890 4.183 2.603 0.00753 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Adjusted p values reported -- none method)Detach (automatically) loaded packages (if possible)
try(detach(package:effects))
try(detach(package:colorspace))
try(detach(package:lattice))
try(detach(package:grid))
try(detach(package:car))
try(detach(package:multcomp))
try(detach(package:survival))
try(detach(package:mvtnorm))
try(detach(package:splines))
try(detach(package:TH.data))Get the article source from GitHub
R markdown - markdown - R code - all posts