Assess univariate and multivariate normality
Install required packages
wants <- c("energy", "ICS", "mvtnorm")
has <- wants %in% rownames(installed.packages())
if(any(!has)) install.packages(wants[!has])Univariate normality
QQ-plot
set.seed(123)
DV <- rnorm(20, mean=1.5, sd=3)
qqnorm(DV, pch=20, cex=2)
qqline(DV, col="gray60", lwd=2)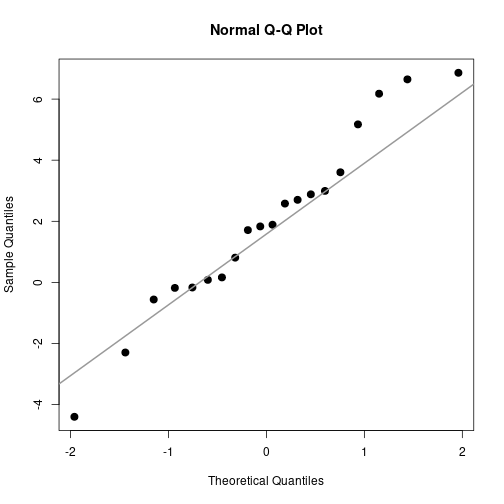
Shapiro-Wilk-test
Composite null hypothesis: any normal distribution
shapiro.test(DV)
Shapiro-Wilk normality test
data: DV
W = 0.9686, p-value = 0.7247Anderson-Darling-test
Composite null hypothesis: any normal distribution
library(DescTools)
AndersonDarlingTest(DV)
Anderson-Darling test of goodness-of-fit
Null hypothesis: uniform distribution
data: DV
An = Inf, p-value = 3e-05Cramer-von-Mises-test
Composite null hypothesis: any normal distribution
library(DescTools)
CramerVonMisesTest(DV)
Cramer-von Mises normality test
data: DV
W = 0.038, p-value = 0.7014Shapiro-Francia-test
Composite null hypothesis: any normal distribution
library(DescTools)
ShapiroFranciaTest(DV)
Shapiro-Francia normality test
data: DV
W = 0.9701, p-value = 0.6667Jarque-Bera-test
Composite null hypothesis: any normal distribution
library(DescTools)
JarqueBeraTest(DV)
Robust Jarque Bera Test
data: DV
X-squared = 0.0183, df = 2, p-value = 0.9909Kolmogorov-Smirnov-test
Exact null hypothesis: fully specified normal distribution
ks.test(DV, "pnorm", mean=1, sd=2, alternative="two.sided")
One-sample Kolmogorov-Smirnov test
data: DV
D = 0.239, p-value = 0.1726
alternative hypothesis: two-sidedLilliefors-test
Composite null hypothesis: any normal distribution
library(DescTools)
LillieTest(DV)
Lilliefors (Kolmogorov-Smirnov) normality test
data: DV
D = 0.1071, p-value = 0.7925Pearson \(\chi^{2}\)-test
Tests weaker null hypothesis (any distribution with the same probabilities for the given class intervals).
Wrong: pearson.test() does not use grouped ML-estimate or maximum \(\chi^{2}\)-estimate
library(DescTools)
PearsonTest(DV, n.classes=6, adjust=TRUE)
Pearson chi-square normality test
data: DV
P = 3.4, p-value = 0.334Multivariate normality
Energy-test
mu <- c(2, 4, 5)
Sigma <- matrix(c(4,2,-3, 2,16,-1, -3,-1,9), byrow=TRUE, ncol=3)
library(mvtnorm)
X <- rmvnorm(100, mu, Sigma)library(energy) # for mvnorm.etest()
mvnorm.etest(X)
Energy test of multivariate normality: estimated parameters
data: x, sample size 100, dimension 3, replicates 999
E-statistic = 1.109, p-value = 0.03704Kurtosis- and skew-test
Kurtosis-test
library(ICS)
mvnorm.kur.test(X)
Multivariate Normality Test Based on Kurtosis
data: X
W = 11.0848, w1 = 1.12, df1 = 5.00, w2 = 1.60, df2 = 1.00, p-value
= 0.1613Skew-test
library(ICS)
X <- rmvnorm(100, c(2, 4, 5))
mvnorm.skew.test(X)
Multivariate Normality Test Based on Skewness
data: X
U = 2.6575, df = 3, p-value = 0.4475Detach (automatically) loaded packages (if possible)
try(detach(package:DescTools))
try(detach(package:energy))
try(detach(package:ICS))
try(detach(package:mvtnorm))
try(detach(package:survey))
try(detach(package:CompQuadForm))Get the article source from GitHub
R markdown - markdown - R code - all posts